Science
Moonwalk’s platform discovers and validates novel, causal targets in obesity
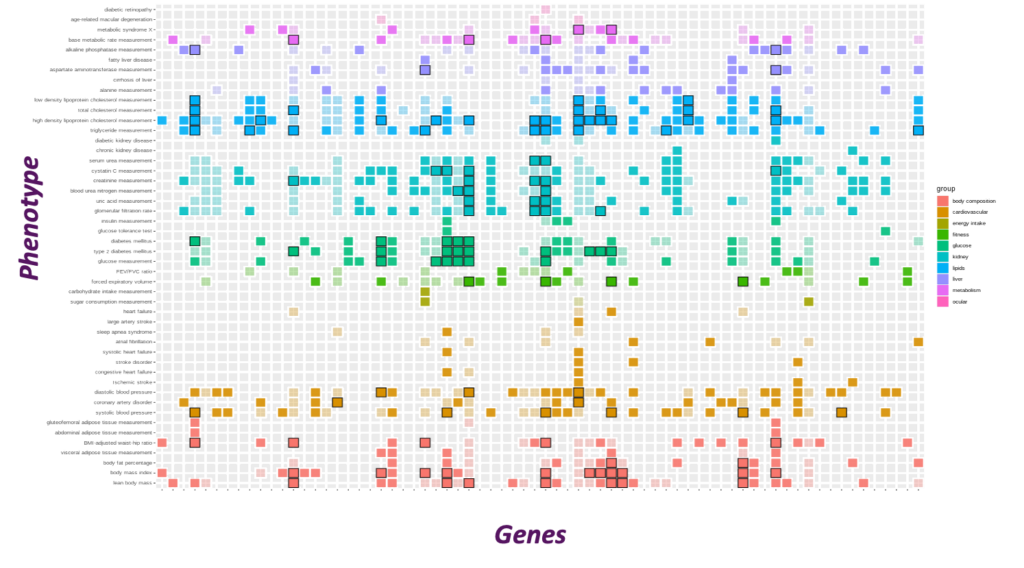
Comprehensive metabolic trait genetic analysis
Genes with human genetic support were identified through mapping genome-wide association study loci across a comprehensive panel of metabolic traits. Approaches used to map variants to genes include spatial proximity, colocalization with expression quantitative trait loci, identification of protein coding variants, and cross referencing with gene burden tests.
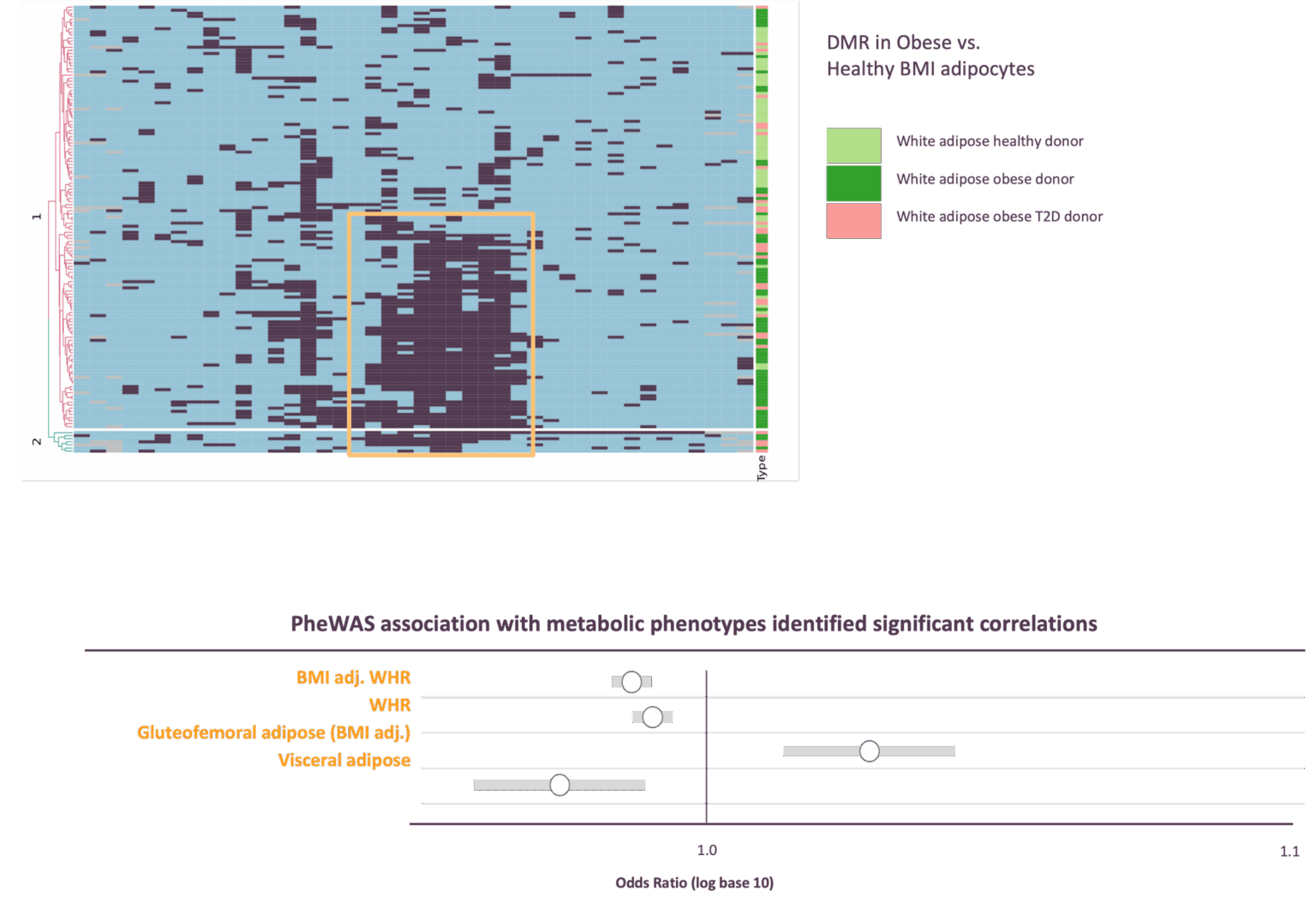
Comprehensive overlay of Epigenetic view of adipose cell states through genome wide methylation
Primary human white adipose tissue-derived adipocytes and preadipocytes from healthy, overweight and obese BMI donors were subjected to whole-genome DNA methylation analysis. Brown adipocytes cultured from human supraclavicular brown adipose tissue depots were also analyzed. Differentially methylated regions were identified and catalogued.
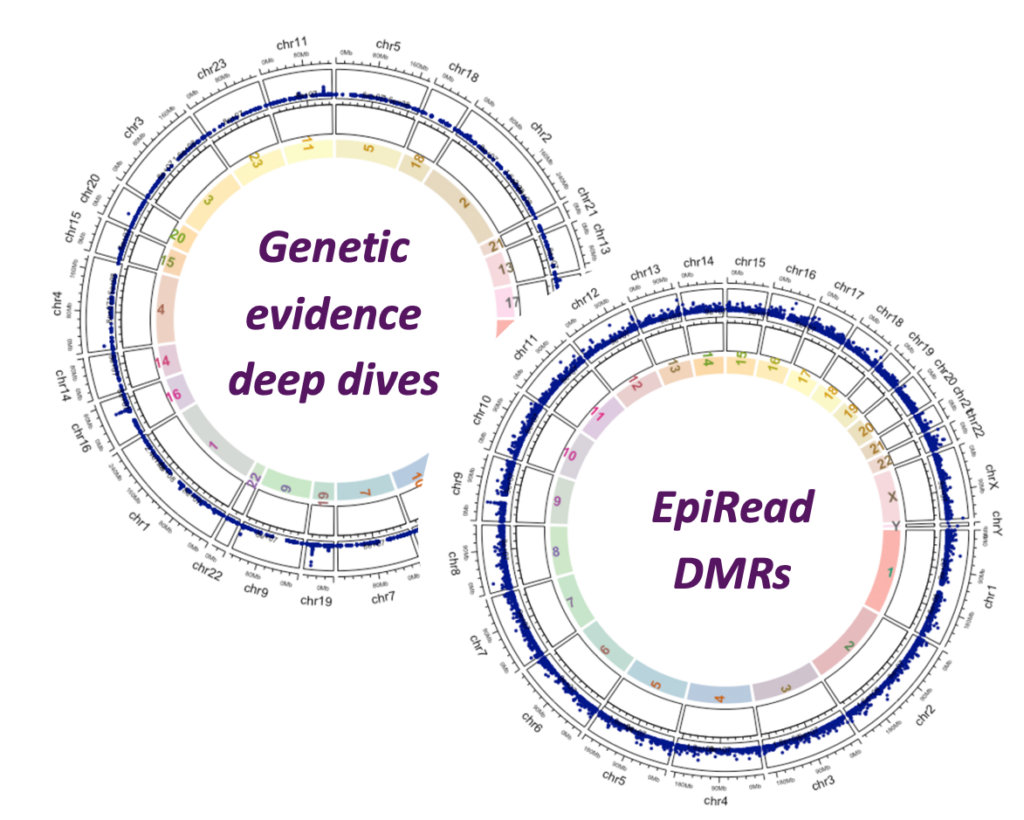
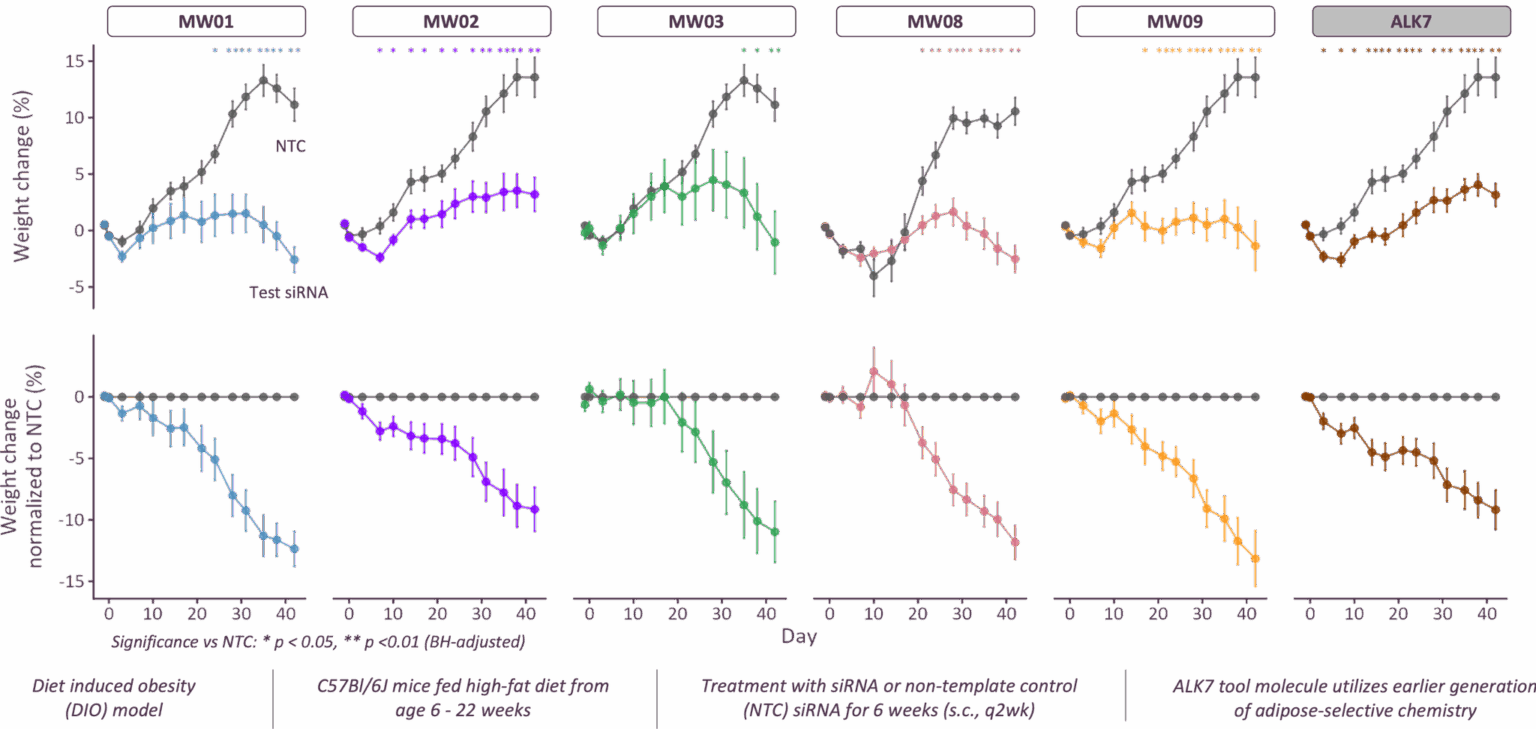
Novel target prediction at the intersection and validation in disease relevant models to select causal targets
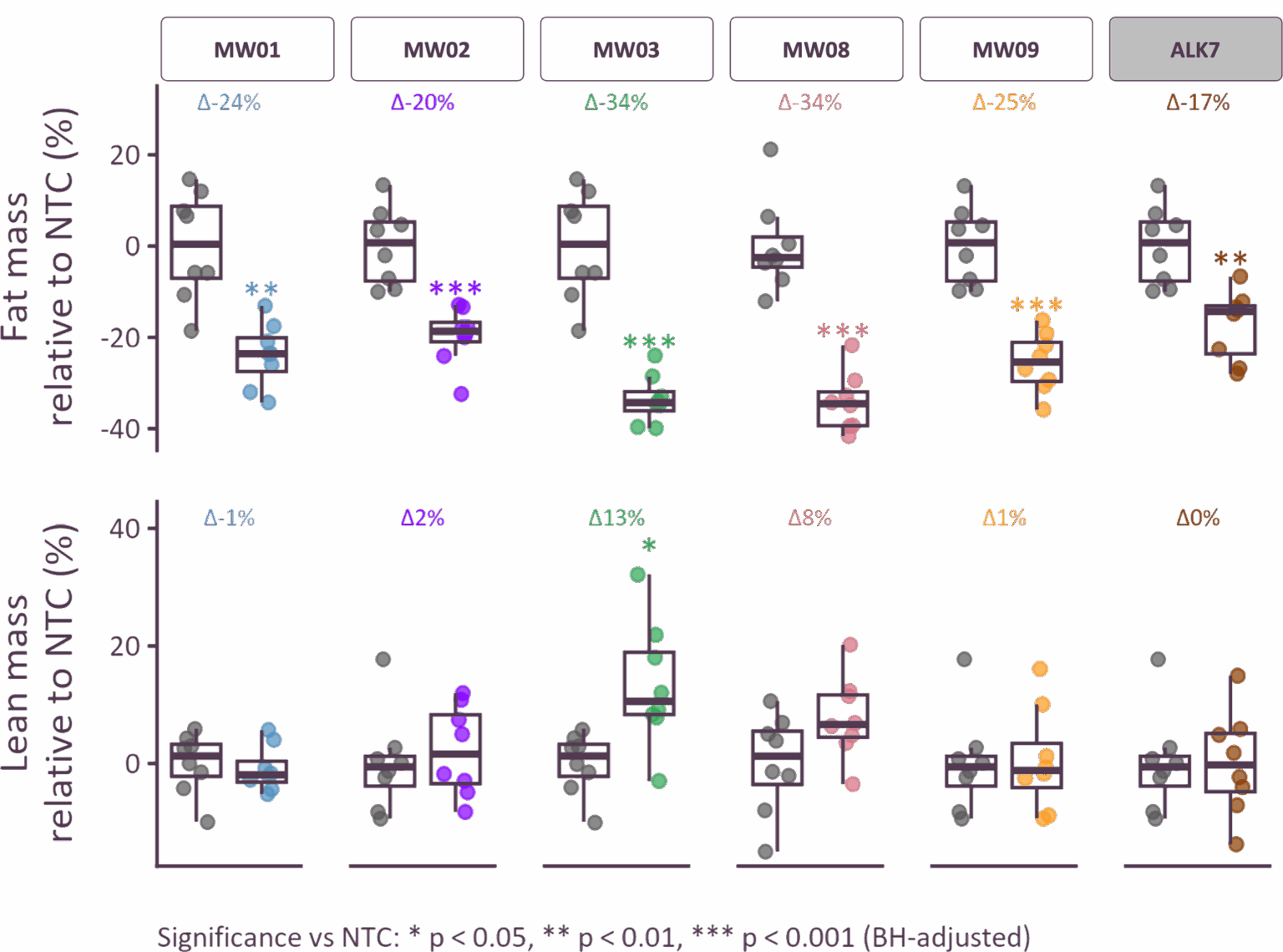
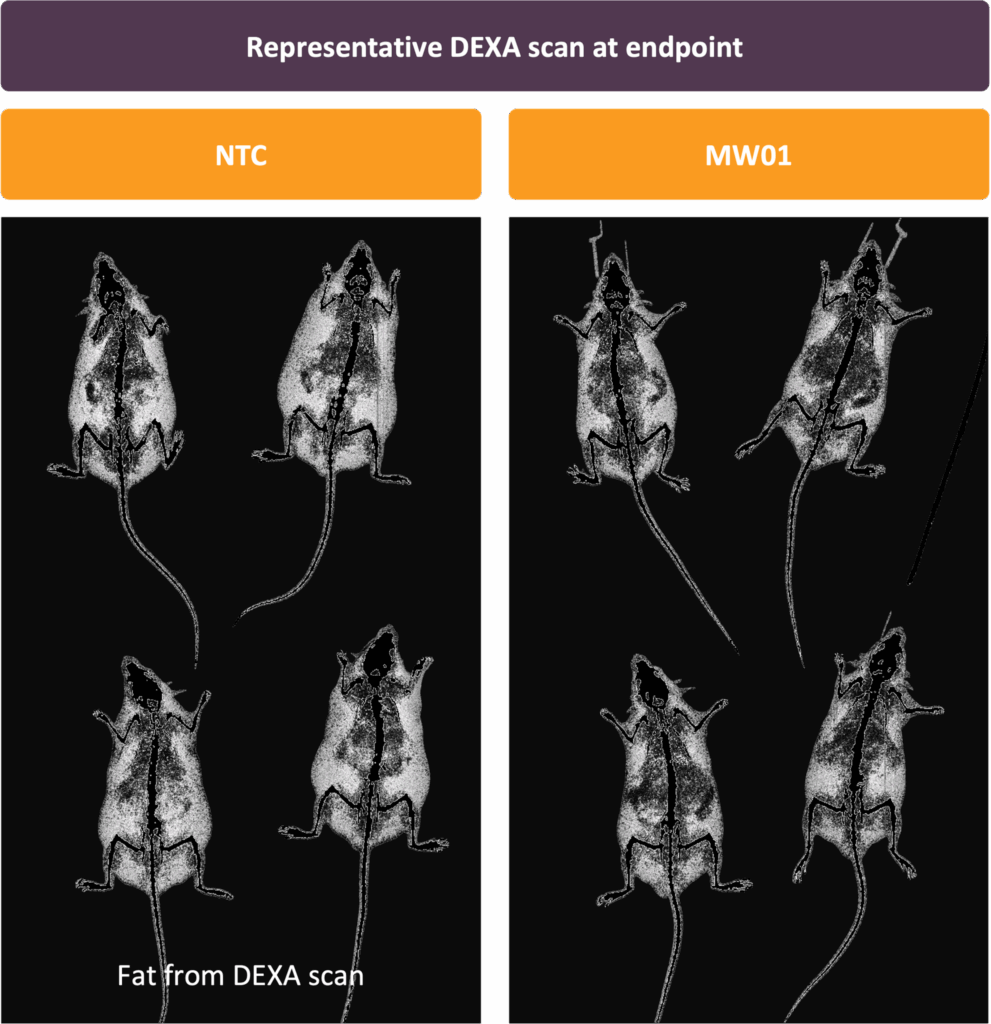
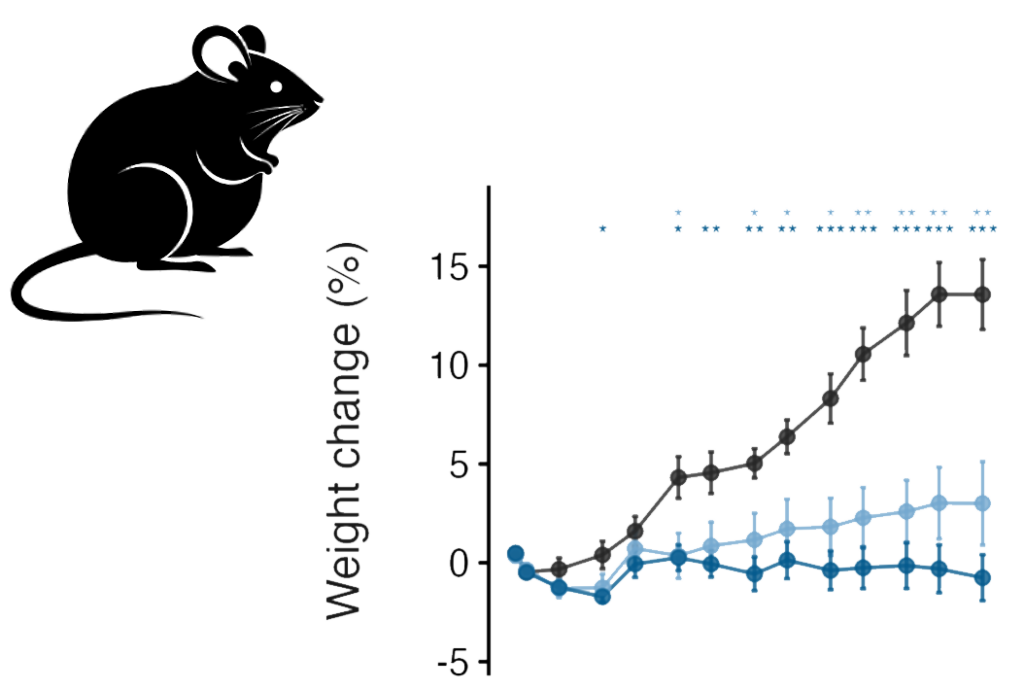
Screening campaigns to identify high-efficiency siRNA sequences to target these genes in rodents. Modification and conjugation of siRNA sequences to mediate adipose-tissue targeting was performed, and adipose-targeted siRNA constructs were tested for in vivo silencing efficacy and therapeutic activity in diet-induced obesity (DIO) models.
Treating obesity by directly targeting novel adipose biology
Moonwalk’s discovery platform resulted in novel adipose targets from deep genetic and epigenetic associations, and human sample testing
Primary human white adipose tissue-derived adipocytes and preadipocytes from healthy, overweight and obese BMI donors were subjected to whole-genome DNA methylation analysis. Brown adipocytes cultured from human supraclavicular brown adipose tissue depots were also analyzed. Differentially methylated regions were identified and catalogued.